How to download NCBI SRA (Sequence Read Archive) data through Google Cloud (GCP)?
In some cases, you might need to download the orignal file from SRA, instead of downloading the data in *.sra format.
- You need the
*.bamor.*h5format for 3rd generation sequencing data. - You would like to have the orignal read name for the
*.fastqformat.
NCBI do not provide download link for the orignal files, but already stored the files on both Amazon Cloud (AWS) and Google Cloud. You can find the data link in the Run Browser page.
Take dataset SRR13168868 as an example, you can have more details on page https://trace.ncbi.nlm.nih.gov/Traces/sra/?run=SRR13168868. And Under Data access tab, there are links startswith “gs://” or “s3://” prefix. The “gs://” one is from Google Cloud (GCP).
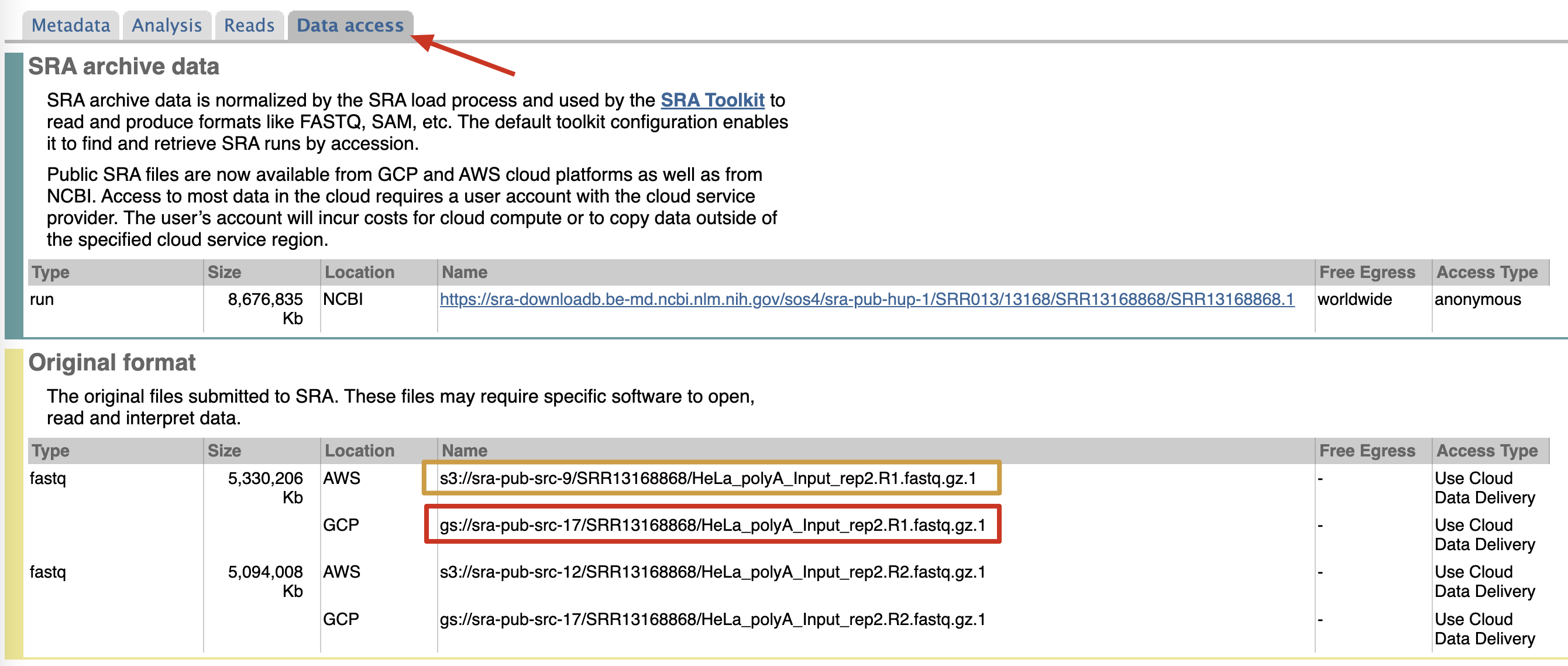
There is an instruction page on NCBI, but it is to easy to follow, and do not provide command line demo. There are some articles about how to download data from AWS, but very few about GCP. So I will only describe GCP.
A google cloud account is needed, and you might also need to complete the payment method.
On the linux server, download and install the
gcloudpackage, with thegsutilcommand.Don’t forget to login the account.
gcloud auth login
If not, you will have an error mesage during the data download.
…serviceusage.services.use access to the Google Cloud Project…
- Creat a key follow the instruction below.
https://cloud.google.com/iam/docs/creating-managing-service-account-keys
And download the key in json format
- You can download the data link by command:
gsutil -m -u project_name cp gs://sra-pub-src-17/SRR13168867/HeLa_polyA_Input_rep1.R1.fastq.gz.1 ./
replace project_name in the command above into your own project name.
Last modified on 2023-11-01